
WIP: Cooking survival data, 5 minute recipes
2026-01-14
Source:vignettes/cooking-survival-data.Rmd
cooking-survival-data.RmdOverview
Simulation of survival data is important for both theoretical and practical work. In a practical setting we might wish to validate that standard errors are valid even in a rather small sample, or validate that a complicated procedure is doing as intended. Therefore it is useful to have simple tools for generating survival data that looks as much as possible like particular data. In a theoretical setting we often are interested in evaluating the finite sample properties of a new procedure in different settings that often are motivated by a specific practical problem. The aim is provide such tools.
Bender et al. in a nice paper discussed how to generate survival data based on the Cox model, and restricted attention to some of the many useful parametric survival models (weibull, exponential). We here use piecewise linear baseline functions that make it easy to simulate data that follows closely the baseline given by the data using semi or nonparametric models. This makes it easy to capture important aspects of the data.
Different survival models can be cooked, and we here give recipes for hazard and cumulative incidence based simulations. More recipes are given in vignette about recurrent events.
- hazard based.
- cumulative incidence.
- recurrent events (see recurrent events vignette).
Hazard based, Cox models
Given a survival time with cumulative hazard , it follows that with (exponential with rate 1), that will have the same distribution as .
This provides the basis for simulations of survival times with a given hazard and is a consequence of this simple calculation
Similarly if given have hazard on Cox form where is a -dimensional regression coefficient and a baseline hazard funcion, then it is useful to observe also that with has the same distribution as given .
Therefore if the inverse of the cumulative hazard can be computed we can generate survival with a specified hazard function. One useful observation is note that for a piecewise linear continuous cumulative hazard on an interval it is easy to compute the inverse.
Further, we can approximate any cumulative hazard with a piecewise linear continous cumulative hazard and then simulate data according to this approximation. Recall that fitting the Cox model to data will give a piecewise constant cumulative hazard and the regression coefficients so with these at hand we can first approximate the piecewise constant “Breslow”-estimator with a linear upper (or lower bound) by simply connecting the values by straight lines.
Delayed entry
If
given
have hazard on Cox form
and we wish to generate data according
to this hazard for those that are alive at time
,
that is draw from the distribution of
given
(all given
), then we note that
with
and with
has the distributiion we are after.
This is again a consequence of a simple calculation
The engine is to simulate data with a given linear cumulative hazard. First generating survival data based on the cumulative hazard cumhaz:j
nsim <- 1000
chaz <- c(0,1,1.5,2,2.1)
breaks <- c(0,10, 20, 30, 40)
cumhaz <- cbind(breaks,chaz)
X <- rbinom(nsim,1,0.5)
beta <- 0.2
rrcox <- exp(X * beta)
pctime <- rchaz(cumhaz,n=nsim)
pctimecox <- rchaz(cumhaz,rrcox)Now looking at a simple cox model
library(mets)
n <- nsim
data(bmt)
bmt$bmi <- rnorm(408)
dcut(bmt) <- gage~age
data <- bmt
cox1 <- phreg(Surv(time,cause==1)~tcell+platelet+age,data=bmt)
dd <- sim.phreg(cox1,n,data=bmt)
dtable(dd,~status)
#>
#> status
#> 0 1
#> 529 471
scox1 <- phreg(Surv(time,status==1)~tcell+platelet+age,data=dd)
cbind(coef(cox1),coef(scox1))
#> [,1] [,2]
#> tcell -0.6517920 -0.4564152
#> platelet -0.5207454 -0.5113844
#> age 0.4083098 0.3860139
par(mfrow=c(1,1))
plot(scox1,col=2); plot(cox1,add=TRUE,col=1)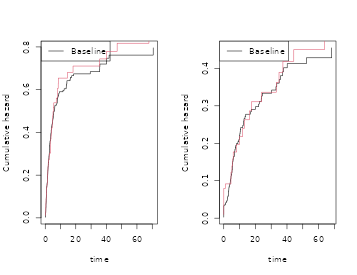
## changing the parameters
cox10 <- cox1
cox10$coef <- c(0,0.4,0.3)
dd <- sim.phreg(cox10,n,data=bmt)
dtable(dd,~status)
#>
#> status
#> 0 1
#> 427 573
scox1 <- phreg(Surv(time,status==1)~tcell+platelet+age,data=dd)
cbind(coef(cox10),coef(scox1))
#> [,1] [,2]
#> tcell 0.0 0.05615982
#> platelet 0.4 0.34930321
#> age 0.3 0.42496872
par(mfrow=c(1,1))
plot(scox1,col=2); plot(cox10,add=TRUE,col=1)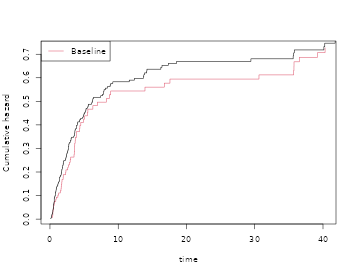
Multiple Cox models for cause specific hazards can be combined, and we start by drawing the covariates manually, below we just call the sim.phregs function that draws covariates from the data,
data(bmt);
cox1 <- phreg(Surv(time,cause==1)~tcell+platelet,data=bmt)
cox2 <- phreg(Surv(time,cause==2)~tcell+platelet,data=bmt)
X1 <- bmt[,c("tcell","platelet")]
n <- nsim
xid <- sample(1:nrow(X1),n,replace=TRUE)
Z1 <- X1[xid,]
Z2 <- X1[xid,]
rr1 <- exp(as.matrix(Z1) %*% cox1$coef)
rr2 <- exp(as.matrix(Z2) %*% cox2$coef)
d <- rcrisk(cox1$cum,cox2$cum,rr1,rr2)
dd <- cbind(d,Z1)
scox1 <- phreg(Surv(time,status==1)~tcell+platelet,data=dd)
scox2 <- phreg(Surv(time,status==2)~tcell+platelet,data=dd)
par(mfrow=c(1,2))
plot(cox1); plot(scox1,add=TRUE,col=2)
plot(cox2); plot(scox2,add=TRUE,col=2)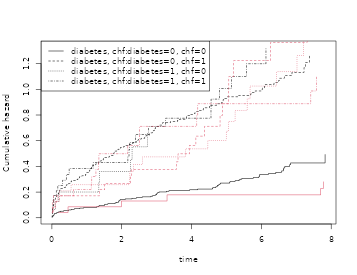
cbind(cox1$coef,scox1$coef,cox2$coef,scox2$coef)
#> [,1] [,2] [,3] [,4]
#> tcell -0.4232606 -0.3727007 0.3991068 0.8167564
#> platelet -0.5654438 -0.5834273 -0.2461474 -0.3190683Now fully nonparametric model with stratified baselines
data(sTRACE)
dtable(sTRACE,~chf+diabetes)
#>
#> diabetes 0 1
#> chf
#> 0 223 16
#> 1 230 31
coxs <- phreg(Surv(time,status==9)~strata(diabetes,chf),data=sTRACE)
strata <- sample(0:3,nsim,replace=TRUE)
simb <- sim.phreg(coxs,nsim,data=NULL,strata=strata)
cc <- phreg(Surv(time,status)~strata(strata),data=simb)
plot(coxs,col=1); plot(cc,add=TRUE,col=2)
simb1 <- sim.phreg(coxs,nsim,data=sTRACE)
cc1 <- phreg(Surv(time,status)~strata(diabetes,chf),data=simb1)
plot(cc1,add=TRUE,col=3)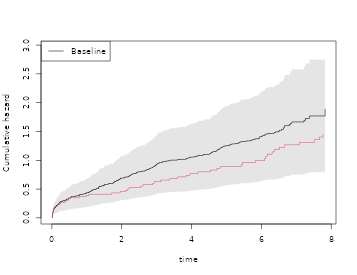
We now fit cause-specific hazard models with 3 causes (censoring as one of them) and generate competing risks data with hazards taken from the fitted Cox models. Here a situation with stratified baselines for some of the models:
## r with phreg
cox0 <- phreg(Surv(time,cause==0)~tcell+platelet,data=bmt)
cox1 <- phreg(Surv(time,cause==1)~tcell+platelet,data=bmt)
cox2 <- phreg(Surv(time,cause==2)~strata(tcell)+platelet,data=bmt)
coxs <- list(cox0,cox1,cox2)
dd <- sim.phregs(coxs,n,data=bmt)
## checking that cause specific hazards are as given, make n larger
scox0 <- phreg(Surv(time,status==1)~tcell+platelet,data=dd)
scox1 <- phreg(Surv(time,status==2)~tcell+platelet,data=dd)
scox2 <- phreg(Surv(time,status==3)~strata(tcell)+platelet,data=dd)
cbind(cox0$coef,scox0$coef)
cbind(cox1$coef,scox1$coef)
cbind(cox2$coef,scox2$coef)
par(mfrow=c(1,3))
plot(cox0); plot(scox0,add=TRUE,col=2);
plot(cox1); plot(scox1,add=TRUE,col=2);
plot(cox2); plot(scox2,add=TRUE,col=2);
########################################
## second example
########################################
cox1 <- phreg(Surv(time,cause==1)~strata(tcell)+platelet,data=bmt)
cox2 <- phreg(Surv(time,cause==2)~tcell+strata(platelet),data=bmt)
coxs <- list(cox1,cox2)
dd <- sim.phregs(coxs,n,data=bmt)
scox1 <- phreg(Surv(time,status==1)~strata(tcell)+platelet,data=dd)
scox2 <- phreg(Surv(time,status==2)~tcell+strata(platelet),data=dd)
cbind(cox1$coef,scox1$coef)
cbind(cox2$coef,scox2$coef)
par(mfrow=c(1,2))
plot(cox1); plot(scox1,add=TRUE);
plot(cox2); plot(scox2,add=TRUE); - sim.phreg for phreg, can deal with strata
- sim.phregs cause specific hazards on phreg form
One more example fully non-parametric
library(mets)
n <- nsim
data(bmt)
bmt$bmi <- rnorm(408)
dcut(bmt) <- gage~age
data <- bmt
cox1 <- phreg(Surv(time,cause==1)~strata(tcell,platelet),data=bmt)
cox2 <- phreg(Surv(time,cause==2)~strata(gage,tcell),data=bmt)
cox3 <- phreg(Surv(time,cause==0)~strata(platelet)+bmi,data=bmt)
coxs <- list(cox1,cox2,cox3)
dd <- sim.phregs(coxs,n,data=bmt,extend=0.002)
dtable(dd,~status)
#>
#> status
#> 0 1 2 3
#> 227 373 232 168
scox1 <- phreg(Surv(time,status==1)~strata(tcell,platelet),data=dd)
scox2 <- phreg(Surv(time,status==2)~strata(gage,tcell),data=dd)
scox3 <- phreg(Surv(time,status==3)~strata(platelet)+bmi,data=dd)
cbind(coef(cox1),coef(scox1), coef(cox2),coef(scox2), coef(cox3),coef(scox3))
#> [,1] [,2]
#> bmi 0.07591238 0.1488615
par(mfrow=c(1,3))
plot(scox1,col=2); plot(cox1,add=TRUE,col=1)
plot(scox2,col=2); plot(cox2,add=TRUE,col=1)
plot(scox3,col=2); plot(cox3,add=TRUE,col=1)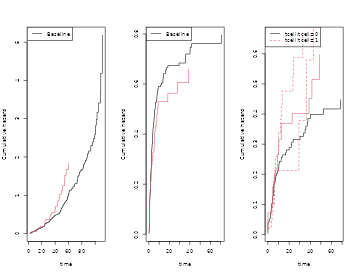
Multistate models: The Illness Death model
Using a hazard based simulation with delayed entry we can then simulate data from for example the general illness-death model. Here the cumulative hazards need to be specified.
We simply give the cumulative hazards for the different transitions to the function simMultistate to simulate data from the model, subsequently we re-estimate the parameters based on the simulated data to validate the procedure.
data(CPH_HPN_CRBSI)
dr <- CPH_HPN_CRBSI$terminal
base1 <- CPH_HPN_CRBSI$crbsi
base4 <- CPH_HPN_CRBSI$mechanical
dr2 <- scalecumhaz(dr,1.5)
cens <- rbind(c(0,0),c(2000,0.5),c(5110,3))
iddata <- simMultistate(nsim,base1,base1,dr,dr2,cens=cens)
dlist(iddata,.~id|id<3,n=0)
#> id: 1
#> entry time status rr death from to start stop
#> 1 0 140.6815 3 1 1 1 3 0 140.6815
#> ------------------------------------------------------------
#> id: 2
#> entry time status rr death from to start stop
#> 2 0.0000 335.4145 2 1 0 1 2 0.0000 335.4145
#> 1001 335.4145 394.7477 1 1 0 2 1 335.4145 394.7477
#> 1634 394.7477 395.6884 0 1 0 1 0 394.7477 395.6884
### estimating rates from simulated data
c0 <- phreg(Surv(start,stop,status==0)~+1,iddata)
c3 <- phreg(Surv(start,stop,status==3)~+strata(from),iddata)
c1 <- phreg(Surv(start,stop,status==1)~+1,subset(iddata,from==2))
c2 <- phreg(Surv(start,stop,status==2)~+1,subset(iddata,from==1))
###
par(mfrow=c(2,2))
plot(c0)
lines(cens,col=2)
plot(c3,main="rates 1-> 3 , 2->3")
lines(dr,col=1,lwd=2)
lines(dr2,col=2,lwd=2)
###
plot(c1,main="rate 1->2")
lines(base1,lwd=2)
###
plot(c2,main="rate 2->1")
lines(base1,lwd=2)
Cumulative incidence
In this section we discuss how to simulate competing risks data that have a specfied cumulative incidence function. We consider for simplicity a competing risks model with two causes and denote the cumulative incidence curves as and . Here given some covariate .
To generate data with the required cumulative incidence functions a simple approach is to first figure out if the subject dies and then from what cause, then finally draw the survival time according to the conditional distribution.
For simplicity we consider survival times in a fixed interval , and first flip a coin with and probabilities to decide if the subject is a survivor or dies. Then if subject dies we then flip a coin with probabilities and to decide if it is a cause , , or a cause 2, . Finally we draw the survival time using the cumulative incidence distribution. The timing of a cause event is thus with and is a uniform.
Then indeed for .
We again note and use that if and are piecewise linear continuous functions then the inverse is easy to compute.
Cumulative incidence I
We here simulate two causes of death with two binary covarites of logistic type and here enforcing the sum condition or not
The baselines are given as where and are postive constants, and here .
To simulate the survival time we use a piecwise linear approximation of the cumulative incidence functions and will thus depends on some grid for linear approximation. Our linear approximation can be made arbitrarily close to any specific smooth cumulative incidence function.
library(mets)
nsim <- 100
rho1 <- 0.4; rho2 <- 2
beta <- c(0.3,-0.3,-0.3,0.3)
dats <- simul.cifs(nsim,rho1,rho2,beta,rc=0.5,depcens=0,type="logistic")
par(mfrow=c(1,2))
# Fitting regression model with CIF logistic-link
cif1 <- cifreg(Event(time,status)~Z1+Z2,dats)
summary(cif1)
#>
#> n events
#> 100 19
#>
#> 100 clusters
#> coeffients:
#> Estimate S.E. dU^-1/2 P-value
#> Z1 -0.097975 0.252782 0.232452 0.6983
#> Z2 0.146136 0.502409 0.464850 0.7711
#>
#> exp(coeffients):
#> Estimate 2.5% 97.5%
#> Z1 0.90667 0.55244 1.4881
#> Z2 1.15735 0.43233 3.0983
plot(cif1)
lines(attr(dats,"Lam1"))
dats <- simul.cifs(nsim,rho1,rho2,beta,rc=0.5,depcens=0,type="cloglog")
ciff <- cifregFG(Event(time,status)~Z1+Z2,dats)
summary(ciff)
#>
#> n events
#> 100 20
#>
#> 100 clusters
#> coeffients:
#> Estimate S.E. dU^-1/2 P-value
#> Z1 0.24189 0.22915 0.23459 0.2912
#> Z2 0.65704 0.46627 0.46940 0.1588
#>
#> exp(coeffients):
#> Estimate 2.5% 97.5%
#> Z1 1.27365 0.81283 1.9957
#> Z2 1.92907 0.77349 4.8110
plot(ciff)
lines(attr(dats,"Lam1"))
We can also use the parameters based on fitted models
data(bmt)
################################################################
# simulating several causes with specific cumulatives
################################################################
## two logistic link models
cif1 <- cifreg(Event(time,cause)~tcell+age,data=bmt,cause=1)
cif2 <- cifreg(Event(time,cause)~tcell+age,data=bmt,cause=2)
dd <- sim.cifs(list(cif1,cif2),nsim,data=bmt)
## still logistic link
scif1 <- cifreg(Event(time,cause)~tcell+age,data=dd,cause=1)
## 2nd cause not on logistic form due to restriction
scif2 <- cifreg(Event(time,cause)~tcell+age,data=dd,cause=2)
cbind(cif1$coef,scif1$coef)
#> [,1] [,2]
#> tcell -0.7966937 0.2319643
#> age 0.4164386 0.7074909
cbind(cif2$coef,scif2$coef)
#> [,1] [,2]
#> tcell 0.66688269 -0.6246131
#> age -0.03248603 -0.5207753
par(mfrow=c(1,2))
plot(cif1); plot(scif1,add=TRUE,col=2)
plot(cif2); plot(scif2,add=TRUE,col=2)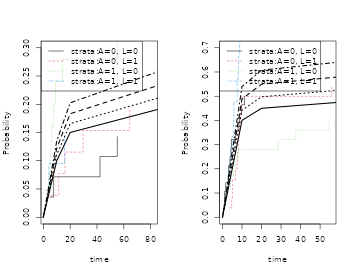
CIF Delayed entry
Now assume that given covariates
and
are two cumulative incidence functions that satistifes the needed
constraints. We wish to generate data that follows these two piecewise
linear cumulative indidence functions with delayed entry at time
.
We should thus generate data that follows the cumulative incidence
functions
and
this can be done according to the
recipe in the previous section.
To be specific (ignoring the
in the formula)
where
is a uniform, will have distribution given by
.
Recurrent events
See also recurrent events vignette
- sim.recurrent can simulate based on the Two-Stage model where the
the
- the rate of the terminal event among survivors in on Cox form
(phreg)
- the rate of the recurrent events among survivors is on Cox form (phreg)
- the rate of the recurrent events is a marginal Ghosh-Lin model (recreg)
- the simulations is based on approximations with piecewise linear models based on a grid.
- the events can be dependent via a frailty random effects (Gamma distributed)
- the rate of the terminal event among survivors in on Cox form
(phreg)
- simRecurrentII, simRecurrent, simRecurrentList
- A frailty Gamma model where the rate of the events and the terminal event are given based on cumulative baselines and relative risk covariate effects. Thus ends up on Cox form given the frailty and covariates.
- simRecurrentList can take multiple recurrent events and multiple causes of death
Two-stage models
data(hfactioncpx12)
hf <- hfactioncpx12
hf$x <- as.numeric(hf$treatment)
n <- 1000
## to fit Cox models
xr <- phreg(Surv(entry,time,status==1)~treatment+cluster(id),data=hf)
dr <- phreg(Surv(entry,time,status==2)~treatment+cluster(id),data=hf)
estimate(xr)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.1534 0.08145 -0.313 0.006286 0.05973
estimate(dr)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.4301 0.1831 -0.7889 -0.07132 0.0188
simcoxcox <- sim.recurrent(xr,dr,n=n,data=hf)
xrs <- phreg(Surv(start,stop,statusD==1)~treatment+cluster(id),data=simcoxcox)
drs <- phreg(Surv(start,stop,statusD==3)~treatment+cluster(id),data=simcoxcox)
estimate(xrs)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.2233 0.07217 -0.3648 -0.08185 0.001974
estimate(drs)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.3901 0.1375 -0.6596 -0.1206 0.004552
par(mfrow=c(1,2))
plot(xrs);
plot(xr,add=TRUE)
###
plot(drs)
plot(dr,add=TRUE)
and a now with Ghosh-Lin and Cox marginals
recGL <- recreg(Event(entry,time,status)~treatment+cluster(id),hf,death.code=2)
estimate(recGL)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.1104 0.07866 -0.2646 0.04376 0.1604
estimate(dr)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.4301 0.1831 -0.7889 -0.07132 0.0188
simglcox <- sim.recurrent(recGL,dr,n=n,data=hf)
simcoxcox <- sim.recurrent(xr,dr,n=n,data=hf)
dtable(simcoxcox,~statusD)
#>
#> statusD
#> 0 1 3
#> 755 2603 245
recGL <- recreg(Event(entry,time,status)~treatment+cluster(id),hf,death.code=2)
simglcox <- sim.recurrent(recGL,dr,n=n,data=hf)
GLs <- recreg(Event(start,stop,statusD)~treatment+cluster(id),data=simglcox,death.code=3)
drs <- phreg(Surv(start,stop,statusD==3)~treatment+cluster(id),data=simglcox)
estimate(GLs)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.1317 0.07259 -0.274 0.0106 0.06968
estimate(drs)
#> Estimate Std.Err 2.5% 97.5% P-value
#> treatment1 -0.46 0.1336 -0.7219 -0.1982 0.0005744
par(mfrow=c(1,2))
plot(GLs);
plot(recGL,add=TRUE)
###
plot(drs)
plot(dr,add=TRUE)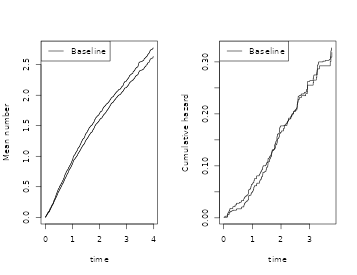
Frailty models
data(CPH_HPN_CRBSI)
dr <- CPH_HPN_CRBSI$terminal
base1 <- CPH_HPN_CRBSI$crbsi
base4 <- CPH_HPN_CRBSI$mechanical
n <- 100
rr <- simRecurrent(n,base1,death.cumhaz=dr)
###
par(mfrow=c(1,3))
showfitsim(causes=1,rr,dr,base1,base1,which=1:2)
rr <- simRecurrentII(n,base1,base4,death.cumhaz=dr)
dtable(rr,~death+status)
#>
#> status 0 1 2
#> death
#> 0 11 266 25
#> 1 89 0 0
showfitsim(causes=2,rr,dr,base1,base4,which=1:2)
cumhaz <- list(base1,base1,base4)
drl <- list(dr,base4)
rr <- simRecurrentList(n,cumhaz,death.cumhaz=drl)
dtable(rr,~death+status)
#>
#> status 0 1 2 3
#> death
#> 0 4 179 192 24
#> 1 78 0 0 0
#> 2 18 0 0 0
showfitsimList(rr,cumhaz,drl) 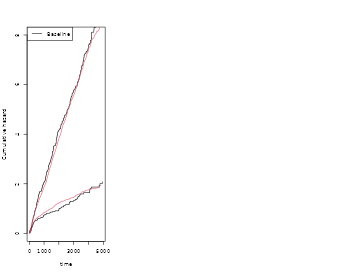

Parametric models
While the semi‑parametric Cox model provides substantial flexibility for simulating survival data, there are situations where a fully parametric simulation model is convenient or preferable. Here we consider a Weibull model parametrized so that the cumulative hazard is given by where is the shape parameter, and the rate parameter. We allow regression on both parameters where and are covariate vectors. Specifically, this opens up for exploring non‑proportional hazards when depends on covariates.
Revisiting the TRACE data example we can compare the predictions from
the Cox and the Weibull-Cox model stratified by chf and
with a proportional hazard effect of age
data(sTRACE, package = "mets")
dat <- sTRACE
cox1 <- phreg(Surv(time, status > 0) ~ strata(chf) + I(age - 67), data = sTRACE)
coxw <- phreg_weibull(Surv(time, status > 0) ~ chf + age,
shape.formula = ~chf,
data = sTRACE
)
coxw
#>
#> - Weibull-Cox model -
#>
#> Call:
#> phreg_weibull(formula = Surv(time, status > 0) ~ chf + age, shape.formula = ~chf,
#> data = sTRACE)
#>
#> log-Likelihood: -684.750499
#>
#> n events obs.time
#> 500 264 2228.481
#>
#> Estimate Std.Err 2.5% 97.5% P-value
#> (Intercept) -5.59626 0.465886 -6.50938 -4.6831 3.070e-33
#> chf 0.83250 0.197629 0.44516 1.2198 2.526e-05
#> age 0.05331 0.006165 0.04123 0.0654 5.241e-18
#> ─────────────
#> s:(Intercept) -0.44096 0.116740 -0.66977 -0.2122 1.585e-04
#> s:chf -0.11794 0.133078 -0.37877 0.1429 3.755e-01
tt <- seq(0, max(sTRACE$time), length.out = 100)
newd <- data.frame(chf = c(1, 0), age=67)
pr <- predict(coxw, newdata = newd, times = tt, type="chaz")
plot(cox1, col = 1)
lines(tt, pr[, 1, 1], lty=2, lwd=2)
lines(tt, pr[, 1, 2], lty = 1, lwd = 2)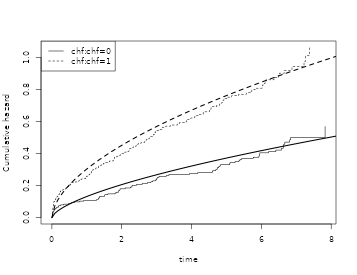
To simulate data we can use the rweibullcox() function.
Note that the stats::rweibull() function gives a different
parametrization where the cumulative hazard is given by
,
i.e., with the same scale parameter but where the scale parameter
is related to the rate parameter we consider by
.
n <- 5000
newd <- mets::dsample(size=n, sTRACE[,c("chf","age")]) # bootstrap covariates
lp <- predict(coxw, newdata=newd, type="lp") # linear-predictors
head(lp)
#> [,1] [,2]
#> X6549 -0.9896742 -0.5589006
#> X6523 -0.5935585 -0.5589006
#> X3742 -1.3657441 -0.5589006
#> X6258 -0.9611517 -0.5589006
#> X79 -2.8312847 -0.4409608
#> X2952 -2.3217722 -0.4409608
## simulate event times
tt <- rweibullcox(nrow(lp), rate = exp(lp[,1]), shape= exp(lp[,2]))
# censoring model
censw <- phreg_weibull(Surv(time, status==0) ~ 1, data=sTRACE)
censpar <- exp(coef(censw))
censtime <- pmin(8, rweibullcox(nrow(lp), censpar[1], censpar[2]))
# combined simulated data
newd <- transform(newd, time=pmin(tt, censtime), status=(tt<=censtime))
head(newd)
#> chf age time status
#> X6549 1 70.791 0.374941 TRUE
#> X6523 1 78.221 3.973340 TRUE
#> X3742 1 63.737 4.330397 TRUE
#> X6258 1 71.326 1.414283 TRUE
#> X79 0 51.863 5.323526 TRUE
#> X2952 0 61.420 6.433051 FALSE
# estimate weibull model on new data
phreg_weibull(Surv(time,status) ~ chf + age, ~chf, data=newd)
#>
#> - Weibull-Cox model -
#>
#> Call:
#> phreg_weibull(formula = Surv(time, status) ~ chf + age, shape.formula = ~chf,
#> data = newd)
#>
#> log-Likelihood: -6682.897120
#>
#> n events obs.time
#> 5000 2622 21530.79
#>
#> Estimate Std.Err 2.5% 97.5% P-value
#> (Intercept) -5.57207 0.154019 -5.87394 -5.27020 1.356e-286
#> chf 0.79395 0.057334 0.68158 0.90632 1.311e-43
#> age 0.05367 0.002115 0.04952 0.05782 5.238e-142
#> ─────────────
#> s:(Intercept) -0.46327 0.032007 -0.52600 -0.40054 1.767e-47
#> s:chf -0.11396 0.038514 -0.18944 -0.03847 3.088e-03All these steps are wrapped in the simulate method:
# simulate(coxw, n = 5, cens.model = NULL, data=newd, var.names = c("time", "status"))
simulate(coxw, nsim = 5)
#> no wmi status chf age sex diabetes time vf
#> X707 707 1.8 TRUE 1 87.175 0 0 0.12940912 0
#> X1157 1157 1.3 FALSE 1 64.074 1 0 7.13699999 0
#> X6628 6628 1.1 TRUE 0 84.825 0 0 0.44463601 0
#> X969 969 0.6 TRUE 1 65.461 1 0 0.05219977 0
#> X4417 4417 1.2 FALSE 0 76.189 1 0 6.01461443 0SessionInfo
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] mets_1.3.9
#>
#> loaded via a namespace (and not attached):
#> [1] cli_3.6.5 knitr_1.51 rlang_1.1.7
#> [4] xfun_0.55 textshaping_1.0.4 jsonlite_2.0.0
#> [7] listenv_0.10.0 future.apply_1.20.1 lava_1.8.2
#> [10] htmltools_0.5.9 ragg_1.5.0 sass_0.4.10
#> [13] rmarkdown_2.30 grid_4.5.2 evaluate_1.0.5
#> [16] jquerylib_0.1.4 fastmap_1.2.0 numDeriv_2016.8-1.1
#> [19] yaml_2.3.12 mvtnorm_1.3-3 lifecycle_1.0.5
#> [22] timereg_2.0.7 compiler_4.5.2 codetools_0.2-20
#> [25] fs_1.6.6 htmlwidgets_1.6.4 Rcpp_1.1.1
#> [28] future_1.68.0 lattice_0.22-7 systemfonts_1.3.1
#> [31] digest_0.6.39 R6_2.6.1 parallelly_1.46.1
#> [34] parallel_4.5.2 splines_4.5.2 Matrix_1.7-4
#> [37] bslib_0.9.0 tools_4.5.2 RcppArmadillo_15.2.3-1
#> [40] globals_0.18.0 survival_3.8-3 pkgdown_2.2.0
#> [43] cachem_1.1.0 desc_1.4.3