
GEE cluster standard errors and predictions for glm objects
Klaus Holst & Thomas Scheike
2026-01-14
Source:vignettes/glm-utility.Rmd
glm-utility.RmdUtility functions for GLM objects
Getting the OR with confidence intervals using the GEE (sandwhich) standard errors
set.seed(100)
library(mets)
data(bmt);
bmt$id <- sample(1:100,408,replace=TRUE)
glm1 <- glm(tcell~platelet+age,bmt,family=binomial)
summaryGLM(glm1)
#> $coef
#> Estimate Std.Err 2.5% 97.5% P-value
#> (Intercept) -2.4371 0.2225 -2.8732 -2.0009 6.481e-28
#> platelet 1.1368 0.3076 0.5340 1.7397 2.189e-04
#> age 0.5927 0.1551 0.2888 0.8966 1.319e-04
#>
#> $or
#> Estimate 2.5% 97.5%
#> (Intercept) 0.08741654 0.05651794 0.1352076
#> platelet 3.11688928 1.70573194 5.6955015
#> age 1.80895115 1.33489115 2.4513641
#>
#> $fout
#> NULL
## GEE robust standard errors
summaryGLM(glm1,id=bmt$id)
#> $coef
#> Estimate Std.Err 2.5% 97.5% P-value
#> (Intercept) -2.4371 0.2157 -2.8599 -2.0142 1.361e-29
#> platelet 1.1368 0.2830 0.5822 1.6914 5.877e-05
#> age 0.5927 0.1434 0.3117 0.8738 3.568e-05
#>
#> $or
#> Estimate 2.5% 97.5%
#> (Intercept) 0.08741654 0.05727471 0.1334211
#> platelet 3.11688928 1.79006045 5.4271903
#> age 1.80895115 1.36575550 2.3959664
#>
#> $fout
#> NULLPredictions also simple
age <- seq(-2,2,by=0.1)
nd <- data.frame(platelet=0,age=seq(-2,2,by=0.1))
pnd <- predictGLM(glm1,nd)
head(pnd$pred)
#> Estimate 2.5% 97.5%
#> p1 0.02601899 0.01115243 0.05951051
#> p2 0.02756409 0.01214068 0.06136414
#> p3 0.02919819 0.01321187 0.06328733
#> p4 0.03092608 0.01437206 0.06528441
#> p5 0.03275278 0.01562757 0.06736019
#> p6 0.03468351 0.01698493 0.06952008
plot(age,pnd$pred[,1],type="l",ylab="predictions",xlab="age",ylim=c(0,0.3))
plotConfRegion(age,pnd$pred[,2:3],col=2)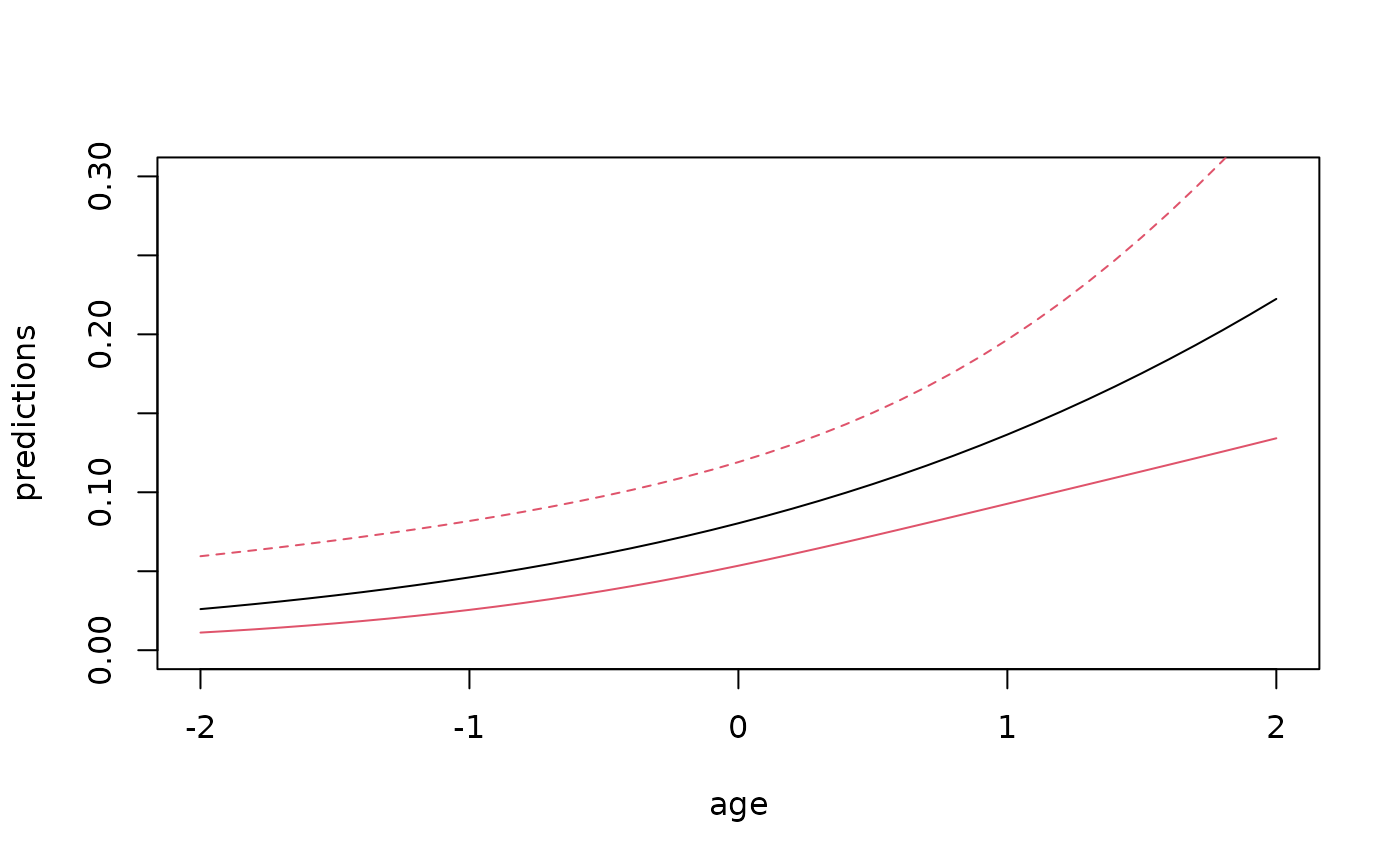
###matlines(age,pnd$pred[,-1],col=2,lty=1)SessionInfo
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] mets_1.3.9
#>
#> loaded via a namespace (and not attached):
#> [1] cli_3.6.5 knitr_1.51 rlang_1.1.7
#> [4] xfun_0.55 textshaping_1.0.4 jsonlite_2.0.0
#> [7] listenv_0.10.0 future.apply_1.20.1 lava_1.8.2
#> [10] htmltools_0.5.9 ragg_1.5.0 sass_0.4.10
#> [13] rmarkdown_2.30 grid_4.5.2 evaluate_1.0.5
#> [16] jquerylib_0.1.4 fastmap_1.2.0 numDeriv_2016.8-1.1
#> [19] yaml_2.3.12 mvtnorm_1.3-3 lifecycle_1.0.5
#> [22] timereg_2.0.7 compiler_4.5.2 codetools_0.2-20
#> [25] fs_1.6.6 htmlwidgets_1.6.4 Rcpp_1.1.1
#> [28] future_1.68.0 lattice_0.22-7 systemfonts_1.3.1
#> [31] digest_0.6.39 R6_2.6.1 parallelly_1.46.1
#> [34] parallel_4.5.2 splines_4.5.2 Matrix_1.7-4
#> [37] bslib_0.9.0 tools_4.5.2 RcppArmadillo_15.2.3-1
#> [40] globals_0.18.0 survival_3.8-3 pkgdown_2.2.0
#> [43] cachem_1.1.0 desc_1.4.3