
Twin models
Klaus Holst & Thomas Scheike
Source:vignettes/quantitative-twin.Rmd
quantitative-twin.RmdThis document provides a brief tutorial to analyzing twin data using
the mets package:
Twin analysis, continuous traits
In the following we examine the heritability of Body Mass Indexkorkeila_bmi_1991hjelmborg_bmi_2008, based on data on self-reported BMI-values from a random sample of 11,411 same-sex twins. First, we will load data
library(mets)
data("twinbmi")
head(twinbmi)
#> tvparnr bmi age gender zyg id num
#> 1 1 26.33289 57.51212 male DZ 1 1
#> 2 1 25.46939 57.51212 male DZ 1 2
#> 3 2 28.65014 56.62696 male MZ 2 1
#> 5 3 28.40909 57.73097 male DZ 3 1
#> 7 4 27.25089 53.68683 male DZ 4 1
#> 8 4 28.07504 53.68683 male DZ 4 2The data is on long format with one subject per row.
-
tvparnr: twin id -
bmi: Body Mass Index () -
age: Age (years) -
gender: Gender factor (male,female) -
zyg: zygosity (MZ, DZ)
We transpose the data allowing us to do pairwise analyses
twinwide <- fast.reshape(twinbmi, id="tvparnr",varying=c("bmi"))
head(twinwide)
#> tvparnr bmi1 age gender zyg id num bmi2
#> 1 1 26.33289 57.51212 male DZ 1 1 25.46939
#> 3 2 28.65014 56.62696 male MZ 2 1 NA
#> 5 3 28.40909 57.73097 male DZ 3 1 NA
#> 7 4 27.25089 53.68683 male DZ 4 1 28.07504
#> 9 5 27.77778 52.55838 male DZ 5 1 NA
#> 11 6 28.04282 52.52231 male DZ 6 1 22.30936Next we plot the association within each zygosity group
We here show the log-transformed data which is slightly more symmetric and more appropiate for the twin analysis (see Figure @ref(fig:scatter1) and @ref(fig:scatter2))
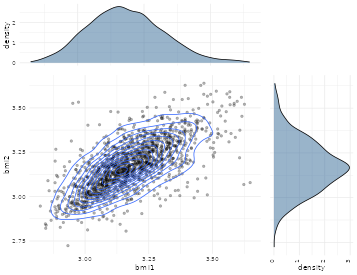
Scatter plot of logarithmic BMI measurements in MZ twins
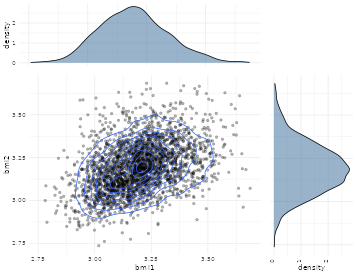
Scatter plot of logarithmic BMI measurements in DZ twins
The plots and raw association measures shows considerable stronger dependence in the MZ twins, thus indicating genetic influence of the trait
cor.test(mz[,1],mz[,2], method="spearman")
#>
#> Spearman's rank correlation rho
#>
#> data: mz[, 1] and mz[, 2]
#> S = 165457624, p-value < 2.2e-16
#> alternative hypothesis: true rho is not equal to 0
#> sample estimates:
#> rho
#> 0.6956209
cor.test(dz[,1],dz[,2], method="spearman")
#>
#> Spearman's rank correlation rho
#>
#> data: dz[, 1] and dz[, 2]
#> S = 2162514570, p-value < 2.2e-16
#> alternative hypothesis: true rho is not equal to 0
#> sample estimates:
#> rho
#> 0.4012686Ńext we examine the marginal distribution (GEE model with working independence)
l0 <- lm(bmi ~ gender + I(age-40), data=twinbmi)
estimate(l0, id=twinbmi$tvparnr)
#> Estimate Std.Err 2.5% 97.5% P-value
#> (Intercept) 23.3687 0.054534 23.2618 23.4756 0.000e+00
#> gendermale 1.4077 0.073216 1.2642 1.5512 2.230e-82
#> I(age - 40) 0.1177 0.004787 0.1083 0.1271 1.499e-133
library("splines")
l1 <- lm(bmi ~ gender*ns(age,3), data=twinbmi)
marg1 <- estimate(l1, id=twinbmi$tvparnr)
dm <- lava::Expand(twinbmi,
bmi=0,
gender=c("male"),
age=seq(33,61,length.out=50))
df <- lava::Expand(twinbmi,
bmi=0,
gender=c("female"),
age=seq(33,61,length.out=50))
plot(marg1, function(p) model.matrix(l1,data=dm)%*%p,
data=dm["age"], ylab="BMI", xlab="Age",
ylim=c(22,26.5))
plot(marg1, function(p) model.matrix(l1,data=df)%*%p,
data=df["age"], col="red", add=TRUE)
legend("bottomright", c("Male","Female"),
col=c("black","red"), lty=1, bty="n")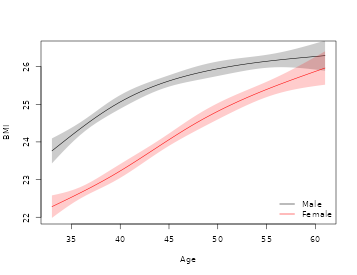
Marginal association between BMI and Age for males and females.
Polygenic model
We can decompose the trait into the following variance components
- : Additive genetic effects of alleles
- : Dominante genetic effects of alleles
- : Shared environmental effects
- : Unique environmental genetic effects
Dissimilarity of MZ twins arises from unshared environmental effects only, $\cor(E_1,E_2)=0$ and
$$\begin{align*} \cor(A_1^{MZ},A_2^{MZ}) = 1, \quad \cor(D_1^{MZ},D_2^{MZ}) = 1, \end{align*}$$
$$\begin{align*} \cor(A_1^{DZ},A_2^{DZ}) = 0.5, \quad \cor(D_1^{DZ},D_2^{DZ}) = 0.25, \end{align*}$$
$$\begin{gather*} \cov(Y_{1},Y_{2}) = \\ \begin{pmatrix} \sigma_A^2 & 2\Phi\sigma_A^2 \\ 2\Phi\sigma_A^2 & \sigma_A^2 \end{pmatrix} + \begin{pmatrix} \sigma_C^2 & \sigma_C^2 \\ \sigma_C^2 & \sigma_C^2 \end{pmatrix} + \begin{pmatrix} \sigma_D^2 & \Delta_{7}\sigma_D^2 \\ \Delta_{7}\sigma_D^2 & \sigma_D^2 \end{pmatrix} + \begin{pmatrix} \sigma_E^2 & 0 \\ 0 & \sigma_E^2 \end{pmatrix} \end{gather*}$$
dd <- na.omit(twinbmi)Saturated model (different marginals in MZ and DZ twins and different marginals for twin 1 and twin 2):
l0 <- twinlm(bmi ~ age+gender, data=dd, DZ="DZ", zyg="zyg", id="tvparnr", type="sat")Different marginals for MZ and DZ twins (but same marginals within a pair)
lf <- twinlm(bmi ~ age+gender, data=dd,DZ="DZ", zyg="zyg", id="tvparnr", type="flex")Same marginals but free correlation with MZ, DZ
lu <- twinlm(bmi ~ age+gender, data=dd, DZ="DZ", zyg="zyg", id="tvparnr", type="eqmarg")
estimate(lu)
#> Estimate Std.Err 2.5% 97.5% P-value
#> bmi.1@1 18.6037 0.251036 18.1116 19.0957 0.000e+00
#> bmi.1~age.1@1 0.1189 0.005635 0.1078 0.1299 9.177e-99
#> bmi.1~gendermale.1@1 1.3848 0.086573 1.2151 1.5544 1.376e-57
#> log(var)@1 2.4424 0.022095 2.3991 2.4857 0.000e+00
#> atanh(rhoMZ)@1 0.7803 0.036249 0.7092 0.8513 9.008e-103
#> atanh(rhoDZ)@2 0.2987 0.020953 0.2576 0.3397 4.288e-46A formal test of genetic effects can be obtained by comparing the MZ and DZ correlation:
estimate(lu,lava::contr(5:6,6))
#> Estimate Std.Err 2.5% 97.5% P-value
#> [atanh(rhoMZ)@1] - [a.... 0.4816 0.04177 0.3997 0.5635 9.431e-31
#>
#> Null Hypothesis:
#> [atanh(rhoMZ)@1] - [atanh(rhoDZ)@2] = 0We also consider the ACE model
ace0 <- twinlm(bmi ~ age+gender, data=dd, DZ="DZ", zyg="zyg", id="tvparnr", type="ace")
summary(ace0)
#> Estimate Std. Error Z value Pr(>|z|)
#> bmi 1.8599e+01 2.5576e-01 72.720 <2e-16
#> sd(A) 2.7270e+00 4.2658e-02 63.927 <2e-16
#> sd(C) 1.7129e-06 3.1064e-01 0.000 1
#> sd(E) 2.0276e+00 3.4787e-02 58.286 <2e-16
#> bmi~age 1.1892e-01 5.6246e-03 21.142 <2e-16
#> bmi~gendermale 1.3846e+00 8.8748e-02 15.601 <2e-16
#>
#> MZ-pairs DZ-pairs
#> 1483 2788
#>
#> Variance decomposition:
#> Estimate 2.5% 97.5%
#> A 0.64399 0.61793 0.67005
#> C 0.00000 0.00000 0.00000
#> E 0.35601 0.32995 0.38207
#>
#>
#> Estimate 2.5% 97.5%
#> Broad-sense heritability 0.64399 0.61793 0.67005
#>
#> Estimate 2.5% 97.5%
#> Correlation within MZ: 0.64399 0.61718 0.66931
#> Correlation within DZ: 0.32200 0.30890 0.33497
#>
#> 'log Lik.' -22019.66 (df=6)
#> AIC: 44051.32
#> BIC: 44089.47Bibliography
[korkeila_bmi_1991] Korkeila, Kaprio, Rissanen & Koskenvuo, Effects of gender and age on the heritability of body mass index, Int J Obes, 15(10), 647-654 (1991). ↩︎
[hjelmborg_bmi_2008] Hjelmborg, Fagnani, Silventoinen, McGue, Korkeila, Christensen, Rissanen & Kaprio, Genetic influences on growth traits of BMI: a longitudinal study of adult twins, Obesity (Silver Spring), 16(4), 847-852 (2008). ↩︎