Cumulative score process residuals for Cox PH regression p-values based on Lin, Wei, Ying resampling.
Usage
# S3 method for class 'phreg'
gof(object, n.sim = 1000, silent = 1, robust = NULL, ...)Examples
library(mets)
data(sTRACE)
m1 <- phreg(Surv(time,status==9)~vf+chf+diabetes,data=sTRACE)
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> vf 7.276731 0.010
#> chf 8.971263 0.074
#> diabetes 3.044404 0.800
par(mfrow=c(1,3))
plot(gg)
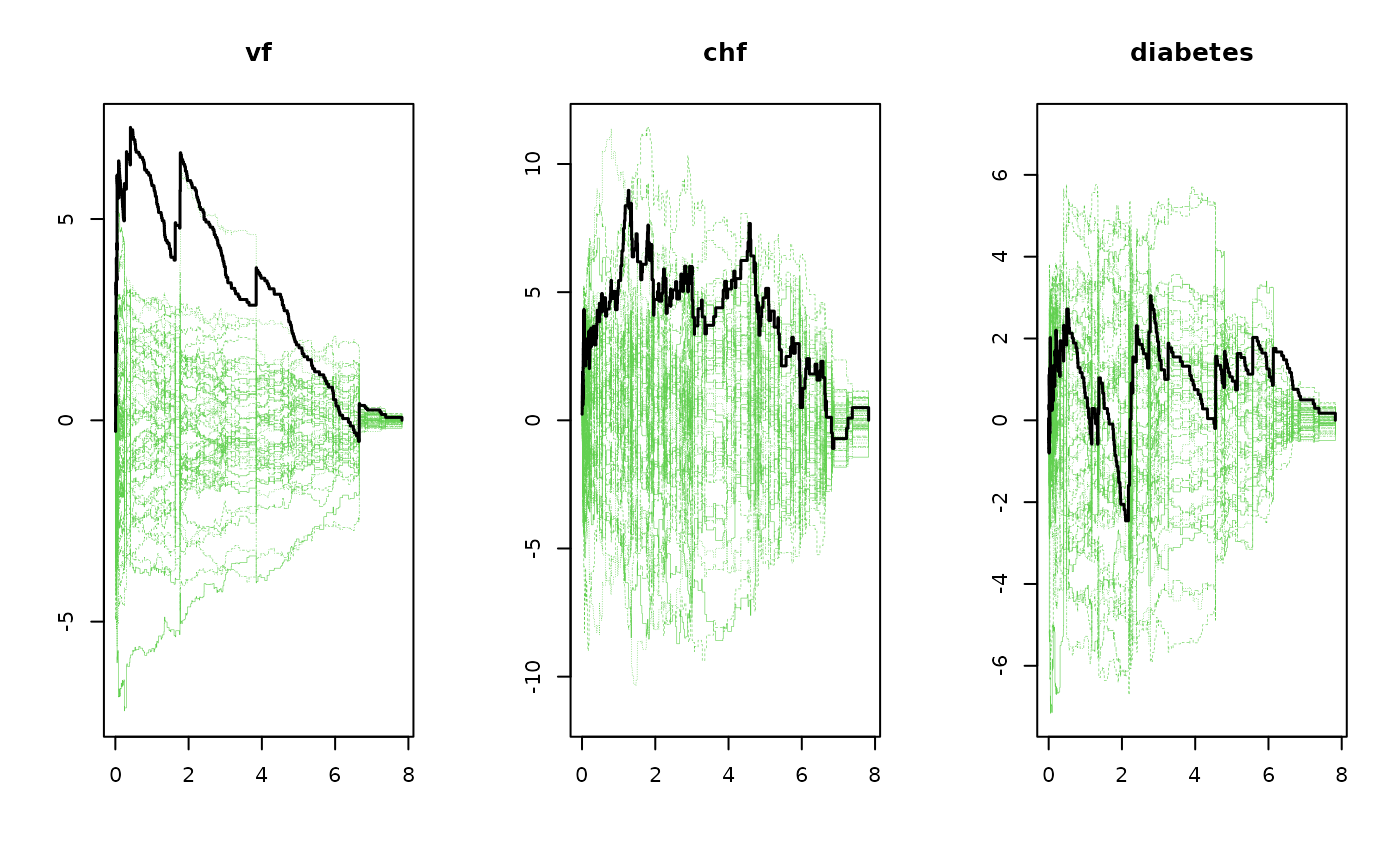 m1 <- phreg(Surv(time,status==9)~strata(vf)+chf+diabetes,data=sTRACE)
## to get Martingale ~ dN based simulations
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> chf 8.036132 0.138
#> diabetes 3.441389 0.697
## to get Martingale robust simulations, specify cluster in call
sTRACE$id <- 1:500
m1 <- phreg(Surv(time,status==9)~vf+chf+diabetes+cluster(id),data=sTRACE)
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> vf 7.276731 0.005
#> chf 8.971263 0.057
#> diabetes 3.044404 0.807
m1 <- phreg(Surv(time,status==9)~strata(vf)+chf+diabetes+cluster(id),data=sTRACE)
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> chf 8.036132 0.141
#> diabetes 3.441389 0.673
m1 <- phreg(Surv(time,status==9)~strata(vf)+chf+diabetes,data=sTRACE)
## to get Martingale ~ dN based simulations
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> chf 8.036132 0.138
#> diabetes 3.441389 0.697
## to get Martingale robust simulations, specify cluster in call
sTRACE$id <- 1:500
m1 <- phreg(Surv(time,status==9)~vf+chf+diabetes+cluster(id),data=sTRACE)
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> vf 7.276731 0.005
#> chf 8.971263 0.057
#> diabetes 3.044404 0.807
m1 <- phreg(Surv(time,status==9)~strata(vf)+chf+diabetes+cluster(id),data=sTRACE)
gg <- gof(m1)
gg
#> Cumulative score process test for Proportionality:
#> Sup|U(t)| pval
#> chf 8.036132 0.141
#> diabetes 3.441389 0.673
