Bivariate Probit model
Usage
biprobit(
x,
data,
id,
rho = ~1,
num = NULL,
strata = NULL,
eqmarg = TRUE,
indep = FALSE,
weights = NULL,
weights.fun = function(x) ifelse(any(x <= 0), 0, max(x)),
randomeffect = FALSE,
vcov = "robust",
pairs.only = FALSE,
allmarg = !is.null(weights),
control = list(trace = 0),
messages = 1,
constrain = NULL,
table = pairs.only,
p = NULL,
...
)Arguments
- x
formula (or vector)
- data
data.frame
- id
The name of the column in the dataset containing the cluster id-variable.
- rho
Formula specifying the regression model for the dependence parameter
- num
Optional name of order variable
- strata
Strata
- eqmarg
If TRUE same marginals are assumed (exchangeable)
- indep
Independence
- weights
Weights
- weights.fun
Function defining the bivariate weight in each cluster
- randomeffect
If TRUE a random effect model is used (otherwise correlation parameter is estimated allowing for both negative and positive dependence)
- vcov
Type of standard errors to be calculated
- pairs.only
Include complete pairs only?
- allmarg
Should all marginal terms be included
- control
Control argument parsed on to the optimization routine. Starting values may be parsed as '
start'.- messages
Control amount of messages shown
- constrain
Vector of parameter constraints (NA where free). Use this to set an offset.
- table
Type of estimation procedure
- p
Parameter vector p in which to evaluate log-Likelihood and score function
- ...
Optional arguments
Examples
data(prt)
prt0 <- subset(prt,country=="Denmark")
a <- biprobit(cancer~1+zyg, ~1+zyg, data=prt0, id="id")
predict(a, newdata=lava::Expand(prt, zyg=c("MZ")))
#> p11 p10 p01 p00 p1 p2 mu1
#> 1 0.005649701 0.01034772 0.01034772 0.9736549 0.01599742 0.01599742 -2.144475
#> mu2 rho parameter zyg
#> 1 -2.144475 0.7547259 1 MZ
## b <- biprobit(cancer~1+zyg, ~1+zyg, data=prt0, id="id",pairs.only=TRUE)
## predict(b,newdata=lava::Expand(prt,zyg=c("MZ","DZ")))
## Reduce Ex.Timings
n <- 2e3
x <- sort(runif(n, -1, 1))
y <- rmvn(n, c(0,0), rho=cbind(tanh(x)))>0
d <- data.frame(y1=y[,1], y2=y[,2], x=x)
dd <- fast.reshape(d)
a <- biprobit(y~1+x,rho=~1+x,data=dd,id="id")
summary(a, mean.contrast=c(1,.5), cor.contrast=c(1,.5))
#>
#> Estimate Std.Err Z p-value
#> (Intercept) -0.0092239 0.0197080 -0.4680 0.6398
#> x 0.0101193 0.0313670 0.3226 0.7470
#> r:(Intercept) -0.0229627 0.0383073 -0.5994 0.5489
#> r:x 0.9894711 0.0692001 14.2987 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> logLik: -2651.885 mean(score^2): 1.394e-05
#> n pairs
#> 4000 2000
#>
#> Contrast:
#> Dependence [(Intercept)] + 0.5[x]
#> Mean [(Intercept)] + 0.5[x]
#>
#> Estimate 2.5% 97.5%
#> Rel.Recur.Risk 1.29171 1.22694 1.35649
#> OR 3.29799 2.55557 4.25610
#> Tetrachoric correlation 0.43963 0.35367 0.51823
#>
#> Concordance 0.32079 0.29429 0.34849
#> Casewise Concordance 0.64371 0.60960 0.67642
#> Marginal 0.49834 0.47536 0.52132
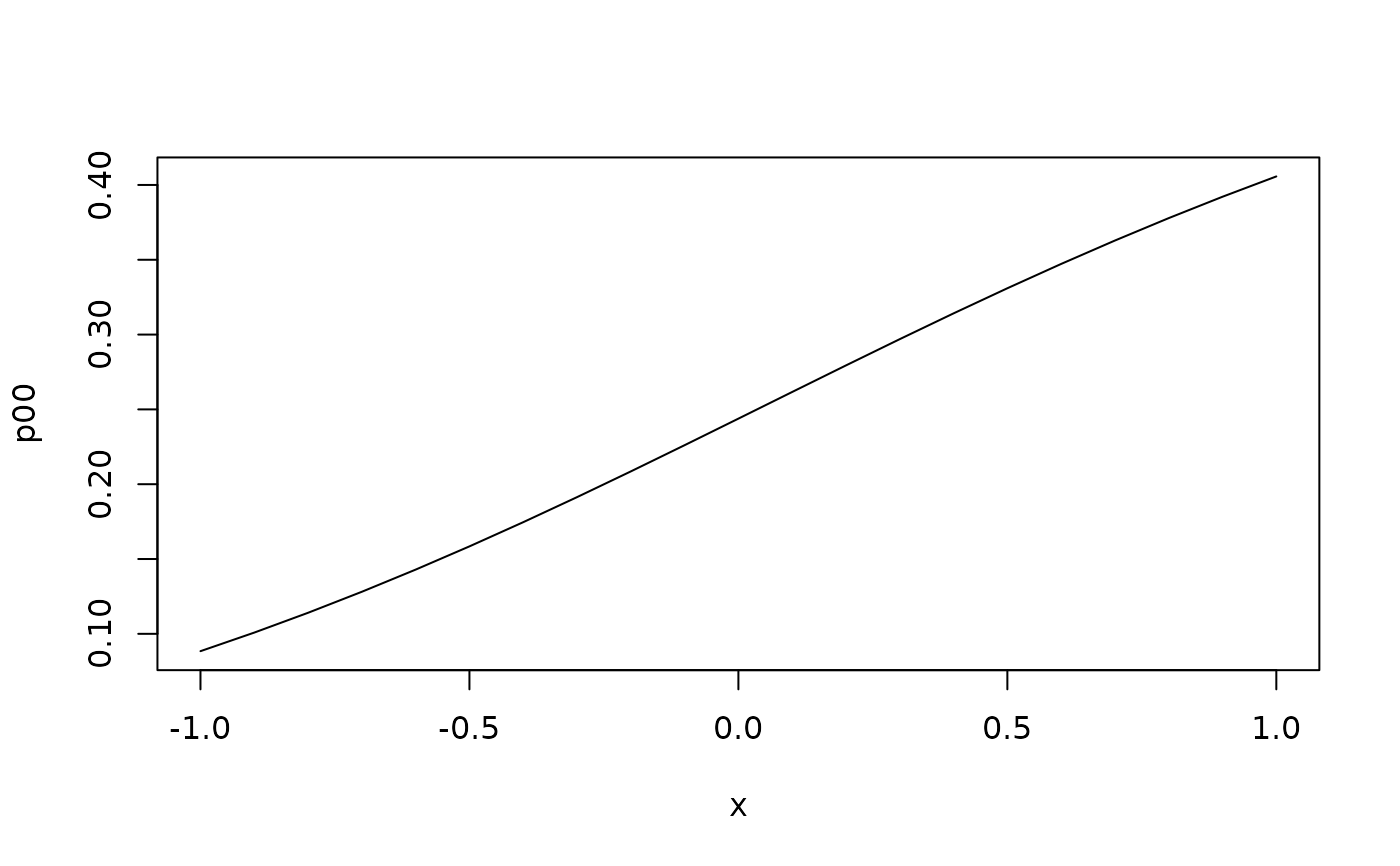
with(predict(a,data.frame(x=seq(-1,1,by=.1))), plot(p00~x,type="l"))
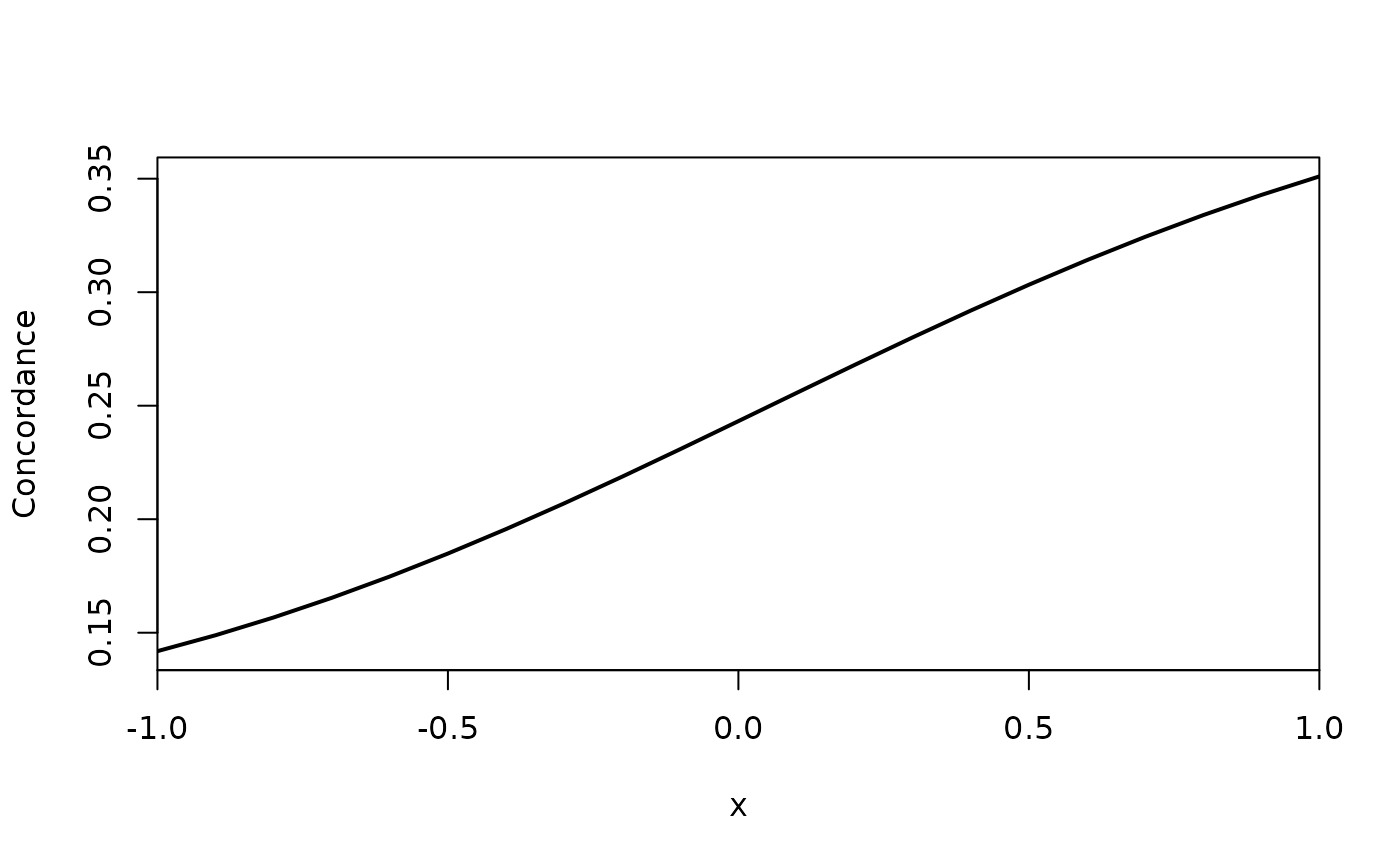
 pp <- predict(a,data.frame(x=seq(-1,1,by=.1)),which=c(1))
plot(pp[,1]~pp$x, type="l", xlab="x", ylab="Concordance", lwd=2, xaxs="i")
lava::confband(pp$x,pp[,2],pp[,3],polygon=TRUE,lty=0,col=lava::Col(1))
pp <- predict(a,data.frame(x=seq(-1,1,by=.1)),which=c(1))
plot(pp[,1]~pp$x, type="l", xlab="x", ylab="Concordance", lwd=2, xaxs="i")
lava::confband(pp$x,pp[,2],pp[,3],polygon=TRUE,lty=0,col=lava::Col(1))
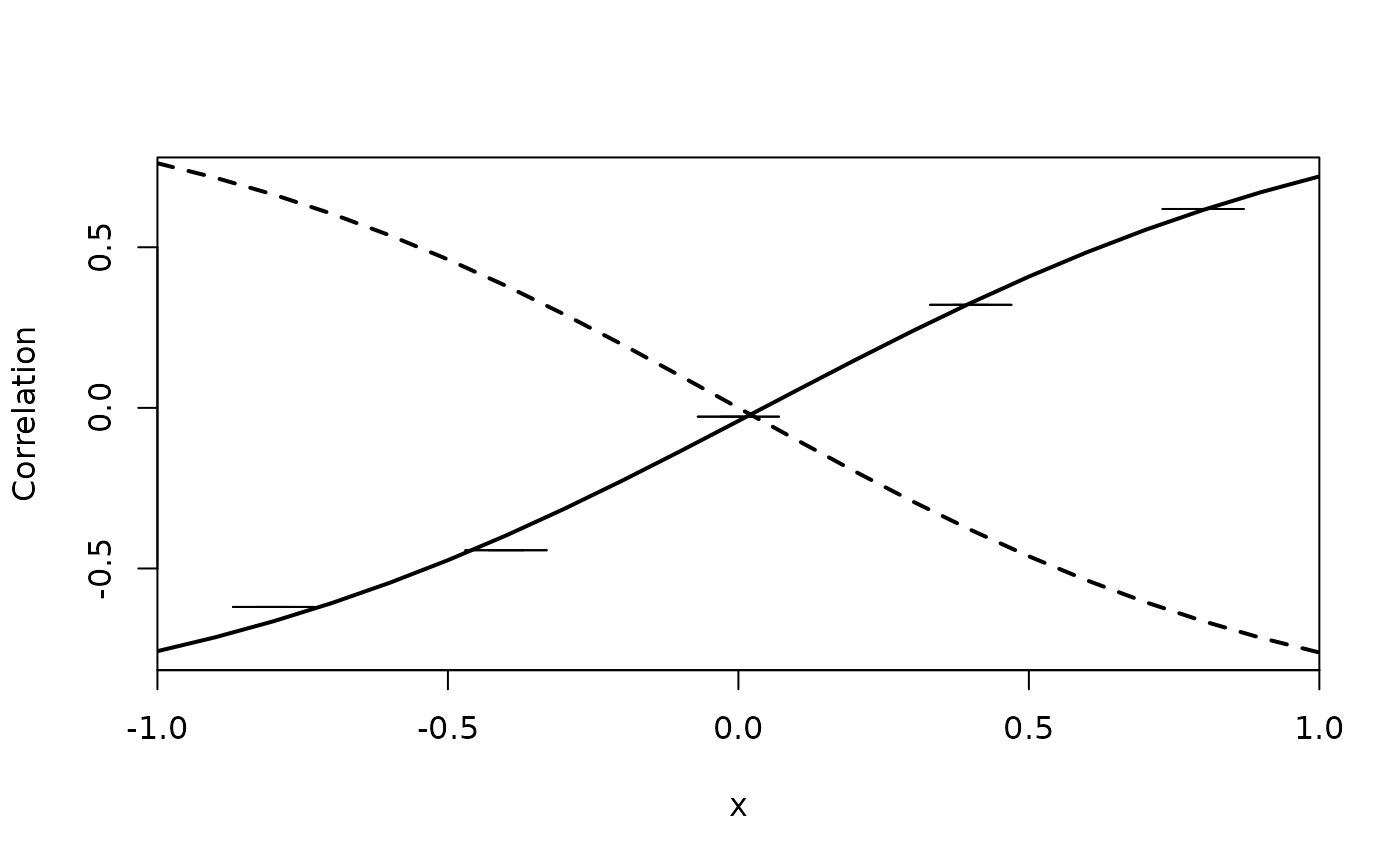
 pp <- predict(a,data.frame(x=seq(-1,1,by=.1)),which=c(9)) ## rho
plot(pp[,1]~pp$x, type="l", xlab="x", ylab="Correlation", lwd=2, xaxs="i")
lava::confband(pp$x,pp[,2],pp[,3],polygon=TRUE,lty=0,col=lava::Col(1))
with(pp, lines(x,tanh(-x),lwd=2,lty=2))
xp <- seq(-1,1,length.out=6); delta <- mean(diff(xp))
a2 <- biprobit(y~1+x,rho=~1+I(cut(x,breaks=xp)),data=dd,id="id")
pp2 <- predict(a2,data.frame(x=xp[-1]-delta/2),which=c(9)) ## rho
lava::confband(pp2$x,pp2[,2],pp2[,3],center=pp2[,1])
pp <- predict(a,data.frame(x=seq(-1,1,by=.1)),which=c(9)) ## rho
plot(pp[,1]~pp$x, type="l", xlab="x", ylab="Correlation", lwd=2, xaxs="i")
lava::confband(pp$x,pp[,2],pp[,3],polygon=TRUE,lty=0,col=lava::Col(1))
with(pp, lines(x,tanh(-x),lwd=2,lty=2))
xp <- seq(-1,1,length.out=6); delta <- mean(diff(xp))
a2 <- biprobit(y~1+x,rho=~1+I(cut(x,breaks=xp)),data=dd,id="id")
pp2 <- predict(a2,data.frame(x=xp[-1]-delta/2),which=c(9)) ## rho
lava::confband(pp2$x,pp2[,2],pp2[,3],center=pp2[,1])
 ## Time
if (FALSE) { # \dontrun{
a <- biprobit.time(cancer~1, rho=~1+zyg, id="id", data=prt, eqmarg=TRUE,
cens.formula=Surv(time,status==0)~1,
breaks=seq(75,100,by=3),fix.censweights=TRUE)
a <- biprobit.time2(cancer~1+zyg, rho=~1+zyg, id="id", data=prt0, eqmarg=TRUE,
cens.formula=Surv(time,status==0)~zyg,
breaks=100)
#a1 <- biprobit.time2(cancer~1, rho=~1, id="id", data=subset(prt0,zyg=="MZ"), eqmarg=TRUE,
# cens.formula=Surv(time,status==0)~1,
# breaks=100,pairs.only=TRUE)
#a2 <- biprobit.time2(cancer~1, rho=~1, id="id", data=subset(prt0,zyg=="DZ"), eqmarg=TRUE,
# cens.formula=Surv(time,status==0)~1,
# breaks=100,pairs.only=TRUE)
} # }
## Time
if (FALSE) { # \dontrun{
a <- biprobit.time(cancer~1, rho=~1+zyg, id="id", data=prt, eqmarg=TRUE,
cens.formula=Surv(time,status==0)~1,
breaks=seq(75,100,by=3),fix.censweights=TRUE)
a <- biprobit.time2(cancer~1+zyg, rho=~1+zyg, id="id", data=prt0, eqmarg=TRUE,
cens.formula=Surv(time,status==0)~zyg,
breaks=100)
#a1 <- biprobit.time2(cancer~1, rho=~1, id="id", data=subset(prt0,zyg=="MZ"), eqmarg=TRUE,
# cens.formula=Surv(time,status==0)~1,
# breaks=100,pairs.only=TRUE)
#a2 <- biprobit.time2(cancer~1, rho=~1, id="id", data=subset(prt0,zyg=="DZ"), eqmarg=TRUE,
# cens.formula=Surv(time,status==0)~1,
# breaks=100,pairs.only=TRUE)
} # }
