
Fast recurrent marginal mean when death is possible
Source:R/recurrent.marginal.R
recurrentMarginal.RdFast Marginal means of recurrent events using the Lin and Ghosh (2000) standard errors. Fitting two models for death and recurent events these are combined to prducte the estimator $$ \int_0^t S(u|x=0) dR(u|x=0) $$ the mean number of recurrent events, here $$ S(u|x=0) $$ is the probability of survival for the baseline group, and $$ dR(u|x=0) $$ is the hazard rate of an event among survivors for the baseline. Here $$ S(u|x=0) $$ is estimated by $$ exp(-\Lambda_d(u|x=0) $$ with $$\Lambda_d(u|x=0) $$ being the cumulative baseline for death.
Details
Assumes no ties in the sense that jump times needs to be unique, this is particularly so for the stratified version.
References
Cook, R. J. and Lawless, J. F. (1997) Marginal analysis of recurrent events and a terminating event. Statist. Med., 16, 911–924. Ghosh and Lin (2002) Nonparametric Analysis of Recurrent events and death, Biometrics, 554–562.
Examples
library(mets)
data(hfactioncpx12)
hf <- hfactioncpx12
hf$x <- as.numeric(hf$treatment)
## to fit non-parametric models with just a baseline
xr <- phreg(Surv(entry,time,status==1)~cluster(id),data=hf)
dr <- phreg(Surv(entry,time,status==2)~cluster(id),data=hf)
par(mfrow=c(1,3))
plot(dr,se=TRUE)
title(main="death")
plot(xr,se=TRUE)
### robust standard errors
rxr <- robust.phreg(xr,fixbeta=1)
plot(rxr,se=TRUE,robust=TRUE,add=TRUE,col=4)
## marginal mean of expected number of recurrent events
## out <- recurrentMarginalPhreg(xr,dr)
## summary(out,times=1:5)
## marginal mean using formula
outN <- recurrentMarginal(Event(entry,time,status)~cluster(id),hf,cause=1,death.code=2)
plot(outN,se=TRUE,col=2,add=TRUE)
summary(outN,times=1:5)
#> [[1]]
#> new.time mean se CI-2.5% CI-97.5% strata
#> 608 1 0.8282358 0.04844543 0.7385251 0.928844 0
#> 1053 2 1.5139493 0.07039884 1.3820710 1.658412 0
#> 1282 3 2.0244982 0.08351867 1.8672476 2.194992 0
#> 1392 4 2.5004732 0.10843166 2.2967320 2.722288 0
#> 1392.1 5 2.5004732 0.10843166 2.2967320 2.722288 0
#>
########################################################################
### with strata ##################################################
########################################################################
out <- recurrentMarginal(Event(entry,time,status)~strata(treatment)+cluster(id),
data=hf,cause=1,death.code=2)
plot(out,se=TRUE,ylab="marginal mean",col=1:2)
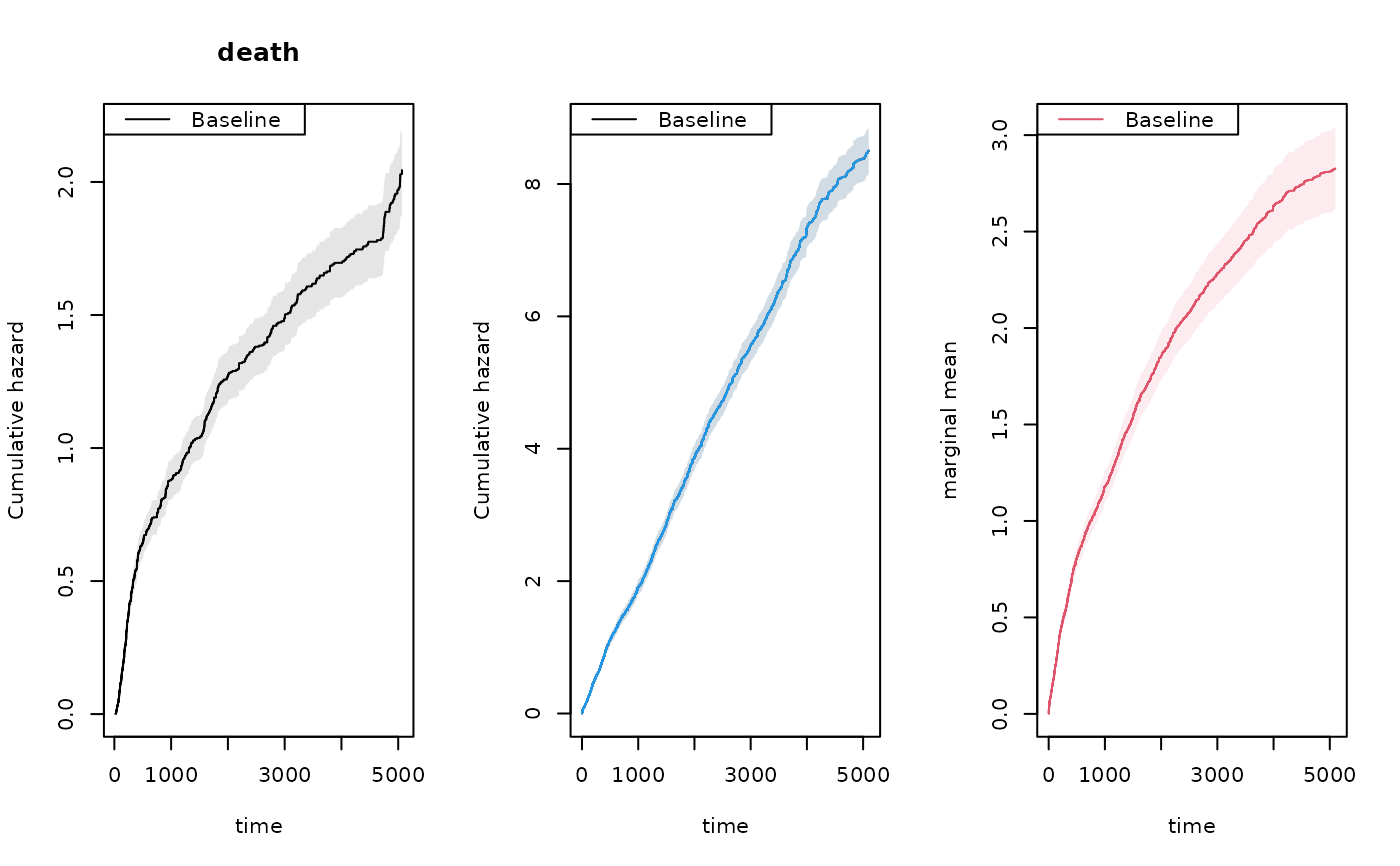 summary(out,times=1:5)
#> [[1]]
#> new.time mean se CI-2.5% CI-97.5% strata
#> 325 1 0.8737156 0.06783343 0.7503858 1.017315 0
#> 555 2 1.5718563 0.09572955 1.3949953 1.771140 0
#> 682 3 2.1184963 0.11385721 1.9066915 2.353829 0
#> 748 4 2.6815219 0.15451005 2.3951619 3.002118 0
#> 748.1 5 2.6815219 0.15451005 2.3951619 3.002118 0
#>
#> [[2]]
#> new.time mean se CI-2.5% CI-97.5% strata
#> 284 1 0.7815557 0.06908585 0.6572305 0.9293989 1
#> 499 2 1.4534055 0.10315606 1.2646561 1.6703258 1
#> 601 3 1.9240624 0.12165771 1.6998008 2.1779119 1
#> 645 4 2.3134997 0.14963892 2.0380418 2.6261880 1
#> 645.1 5 2.3134997 0.14963892 2.0380418 2.6261880 1
#>
summary(out,times=1:5)
#> [[1]]
#> new.time mean se CI-2.5% CI-97.5% strata
#> 325 1 0.8737156 0.06783343 0.7503858 1.017315 0
#> 555 2 1.5718563 0.09572955 1.3949953 1.771140 0
#> 682 3 2.1184963 0.11385721 1.9066915 2.353829 0
#> 748 4 2.6815219 0.15451005 2.3951619 3.002118 0
#> 748.1 5 2.6815219 0.15451005 2.3951619 3.002118 0
#>
#> [[2]]
#> new.time mean se CI-2.5% CI-97.5% strata
#> 284 1 0.7815557 0.06908585 0.6572305 0.9293989 1
#> 499 2 1.4534055 0.10315606 1.2646561 1.6703258 1
#> 601 3 1.9240624 0.12165771 1.6998008 2.1779119 1
#> 645 4 2.3134997 0.14963892 2.0380418 2.6261880 1
#> 645.1 5 2.3134997 0.14963892 2.0380418 2.6261880 1
#>