Simulation of recurrent events data based on cumulative hazards for event and death process
Source:R/recurrent.marginal.R
simRecurrent.RdSimulation of recurrent events data based on cumulative hazards for event and death process
Examples
########################################
## getting some rates to mimick
########################################
library(mets)
data(CPH_HPN_CRBSI)
dr <- CPH_HPN_CRBSI$terminal
base1 <- CPH_HPN_CRBSI$crbsi
base4 <- CPH_HPN_CRBSI$mechanical
######################################################################
### simulating simple model that mimicks data
######################################################################
rr <- simRecurrent(5,base1)
dlist(rr,.~id,n=0)
#> id: 1
#> entry time status dtime fdeath death start stop
#> 1 0.0000 479.9688 1 5110 0 0 0.0000 479.9688
#> 6 479.9688 772.2568 1 5110 0 0 479.9688 772.2568
#> 11 772.2568 957.6359 1 5110 0 0 772.2568 957.6359
#> 16 957.6359 1170.7139 1 5110 0 0 957.6359 1170.7139
#> 21 1170.7139 1484.9421 1 5110 0 0 1170.7139 1484.9421
#> 25 1484.9421 1898.2506 1 5110 0 0 1484.9421 1898.2506
#> 29 1898.2506 2573.3859 1 5110 0 0 1898.2506 2573.3859
#> 32 2573.3859 2866.1914 1 5110 0 0 2573.3859 2866.1914
#> 35 2866.1914 3131.7214 1 5110 0 0 2866.1914 3131.7214
#> 37 3131.7214 3328.1125 1 5110 0 0 3131.7214 3328.1125
#> 38 3328.1125 5110.0000 0 5110 0 0 3328.1125 5110.0000
#> ------------------------------------------------------------
#> id: 2
#> entry time status dtime fdeath death start stop
#> 2 0.000 1817.737 1 5110 0 0 0.000 1817.737
#> 7 1817.737 3877.933 1 5110 0 0 1817.737 3877.933
#> 12 3877.933 5109.602 1 5110 0 0 3877.933 5109.602
#> 17 5109.602 5110.000 0 5110 0 0 5109.602 5110.000
#> ------------------------------------------------------------
#> id: 3
#> entry time status dtime fdeath death start stop
#> 3 0.000 1394.052 1 5110 0 0 0.000 1394.052
#> 8 1394.052 1463.023 1 5110 0 0 1394.052 1463.023
#> 13 1463.023 2037.959 1 5110 0 0 1463.023 2037.959
#> 18 2037.959 2125.253 1 5110 0 0 2037.959 2125.253
#> 22 2125.253 2312.194 1 5110 0 0 2125.253 2312.194
#> 26 2312.194 3232.312 1 5110 0 0 2312.194 3232.312
#> 30 3232.312 3511.355 1 5110 0 0 3232.312 3511.355
#> 33 3511.355 5110.000 0 5110 0 0 3511.355 5110.000
#> ------------------------------------------------------------
#> id: 4
#> entry time status dtime fdeath death start stop
#> 4 0.000 1531.245 1 5110 0 0 0.000 1531.245
#> 9 1531.245 1560.042 1 5110 0 0 1531.245 1560.042
#> 14 1560.042 2371.617 1 5110 0 0 1560.042 2371.617
#> 19 2371.617 3111.797 1 5110 0 0 2371.617 3111.797
#> 23 3111.797 3252.093 1 5110 0 0 3111.797 3252.093
#> 27 3252.093 3486.817 1 5110 0 0 3252.093 3486.817
#> 31 3486.817 3980.201 1 5110 0 0 3486.817 3980.201
#> 34 3980.201 4029.447 1 5110 0 0 3980.201 4029.447
#> 36 4029.447 5110.000 0 5110 0 0 4029.447 5110.000
#> ------------------------------------------------------------
#> id: 5
#> entry time status dtime fdeath death start stop
#> 5 0.000 2037.976 1 5110 0 0 0.000 2037.976
#> 10 2037.976 2776.202 1 5110 0 0 2037.976 2776.202
#> 15 2776.202 2838.842 1 5110 0 0 2776.202 2838.842
#> 20 2838.842 3986.594 1 5110 0 0 2838.842 3986.594
#> 24 3986.594 3990.847 1 5110 0 0 3986.594 3990.847
#> 28 3990.847 5110.000 0 5110 0 0 3990.847 5110.000
rr <- simRecurrent(5,base1,death.cumhaz=dr)
dlist(rr,.~id,n=0)
#> id: 1
#> entry time status dtime fdeath death start stop
#> 1 0 63.83977 0 63.83977 1 1 0 63.83977
#> ------------------------------------------------------------
#> id: 2
#> entry time status dtime fdeath death start stop
#> 2 0 174.8384 0 174.8384 1 1 0 174.8384
#> ------------------------------------------------------------
#> id: 3
#> entry time status dtime fdeath death start stop
#> 3 0 265.97 0 265.97 1 1 0 265.97
#> ------------------------------------------------------------
#> id: 4
#> entry time status dtime fdeath death start
#> 4 0.0000000 0.3237961 1 1713.332 1 0 0.0000000
#> 6 0.3237961 855.0953903 1 1713.332 1 0 0.3237961
#> 8 855.0953903 956.5468201 1 1713.332 1 0 855.0953903
#> 10 956.5468201 1713.3321737 0 1713.332 1 1 956.5468201
#> stop
#> 4 0.3237961
#> 6 855.0953903
#> 8 956.5468201
#> 10 1713.3321737
#> ------------------------------------------------------------
#> id: 5
#> entry time status dtime fdeath death start stop
#> 5 0.0000 217.9460 1 752.5034 1 0 0.0000 217.9460
#> 7 217.9460 301.7446 1 752.5034 1 0 217.9460 301.7446
#> 9 301.7446 546.4851 1 752.5034 1 0 301.7446 546.4851
#> 11 546.4851 752.5034 0 752.5034 1 1 546.4851 752.5034
rr <- simRecurrent(100,base1,death.cumhaz=dr)
par(mfrow=c(1,3))
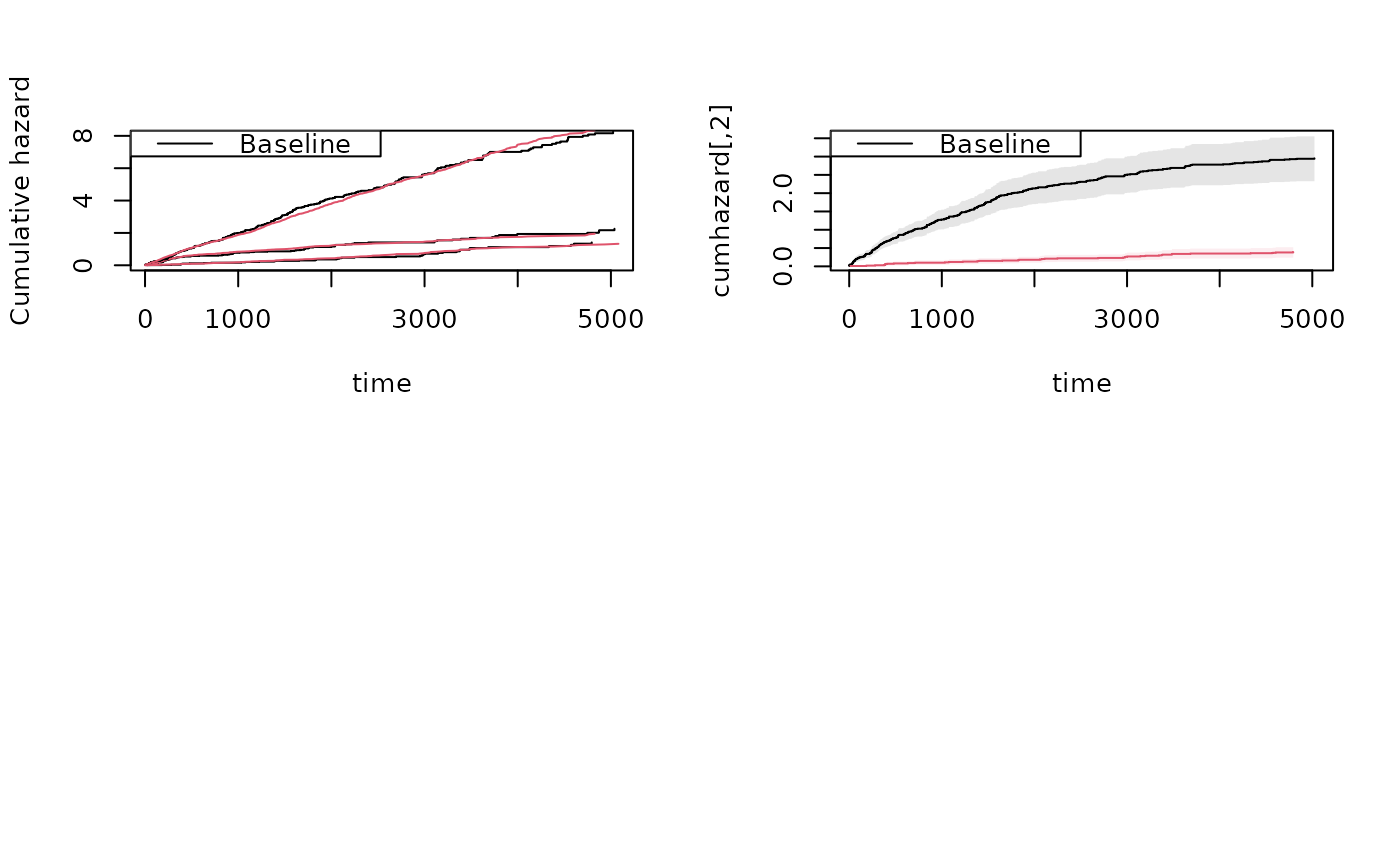
showfitsim(causes=1,rr,dr,base1,base1)
######################################################################
### simulating simple model
### random effect for all causes (Z shared for death and recurrent)
######################################################################
rr <- simRecurrent(100,base1,death.cumhaz=dr,dependence=1,var.z=0.4)
dtable(rr,~death+status)
#>
#> status 0 1
#> death
#> 0 22 237
#> 1 78 0
######################################################################
### now with two event types and second type has same rate as death rate
######################################################################
set.seed(100)
rr <- simRecurrentII(100,base1,base4,death.cumhaz=dr)
dtable(rr,~death+status)
#>
#> status 0 1 2
#> death
#> 0 10 295 39
#> 1 90 0 0
par(mfrow=c(2,2))
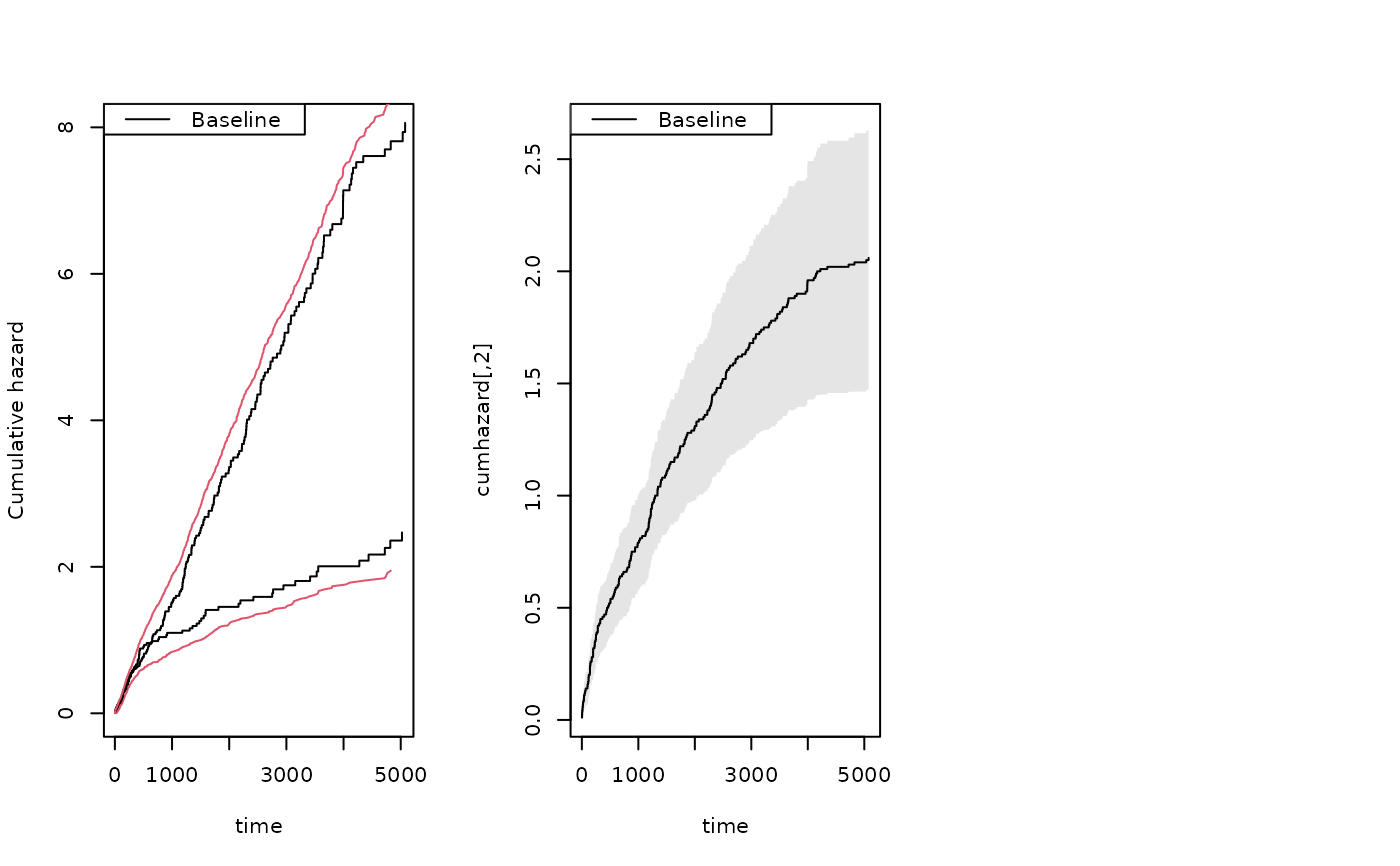 showfitsim(causes=2,rr,dr,base1,base4)
######################################################################
### now with three event types and two causes of death
######################################################################
set.seed(100)
cumhaz <- list(base1,base1,base4)
drl <- list(dr,base4)
rr <- simRecurrentList(100,cumhaz,death.cumhaz=drl,dependence=0)
dtable(rr,~death+status)
#>
#> status 0 1 2 3
#> death
#> 0 4 232 268 33
#> 1 70 0 0 0
#> 2 26 0 0 0
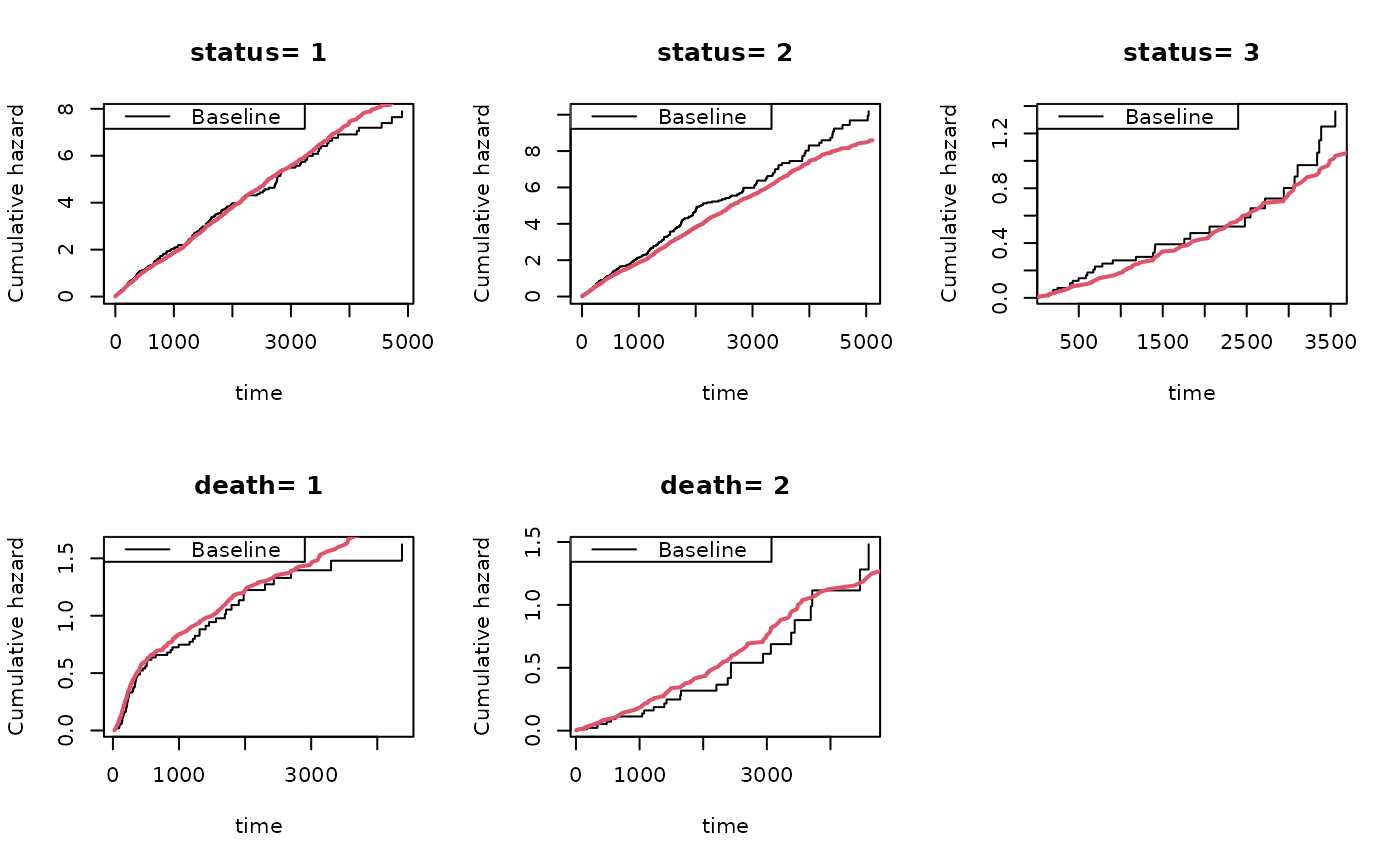
showfitsimList(rr,cumhaz,drl)
showfitsim(causes=2,rr,dr,base1,base4)
######################################################################
### now with three event types and two causes of death
######################################################################
set.seed(100)
cumhaz <- list(base1,base1,base4)
drl <- list(dr,base4)
rr <- simRecurrentList(100,cumhaz,death.cumhaz=drl,dependence=0)
dtable(rr,~death+status)
#>
#> status 0 1 2 3
#> death
#> 0 4 232 268 33
#> 1 70 0 0 0
#> 2 26 0 0 0
showfitsimList(rr,cumhaz,drl)